Aging is a comprehensive and gradual decline in physical fitness, intelligence and reproductive capacity, accompanied by the occurrence of multiple diseases and increased mortality, but the comprehensive understanding of the interaction between genetics, gender and environment in aging and gender determination is still limited. However, aging is not a single-dimensional change. Commonly used aging characteristics are measured based on life span, life history, age-related disease onset and physiological markers. In order to identify genes and pathways related to aging and characterize the genetic and non-genetic determinants of longevity at the population level, the research group of Johan Auwerx at the Ecole Polytechnique Fédérale de Lausanne in Switzerland and the research group of Robert W. Williams at the University of Tennessee in the United States collaborated to publish an article in Science titled Sex- and age-dependent genetics of longevity in a heterogeneous mouse population. Through large-scale screening of mouse longevity genes, they discovered sex-specific and age-specific longevity gene loci, providing important resources for future research on aging, related diseases and longevity.
Aging is a time-dependent functional decline in molecular, cellular, and organ homeostasis influenced by multiple environmental and genetic factors [1]. In addition to metabolic decline, other characteristics such as loss of proteostasis, increased genomic instability, changes in epigenetic marks, and alterations in telomere length are also considered to promote aging. In this work, the authors used an unbiased genetic and molecular cross-species approach to search for drivers of aging. The authors' experiments began with the localization of loci that regulate lifespan variation in male and female untreated control mice from the Interventions Testing Program (ITP) and in multi-year four-way crosses. The ITP Center began testing the effects of dietary and pharmacological interventions on lifespan in 2004 [2]. The authors systematically collected tail biopsies from nearly all mice, allowing for genotyping and subsequent genetic analysis (Figure 1). The authors generated genotype data for 3276 mice, including 2356 control mice and 920 drug-treated mice.
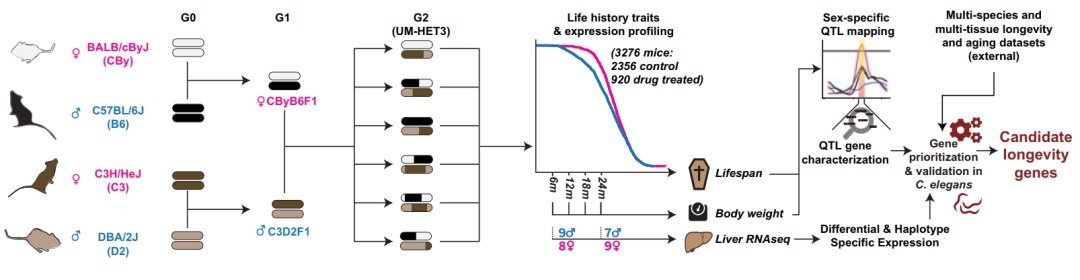
Figure 1 Workflow
The authors found that female mice had a higher median lifespan than males. On the survival curve, male mice had a higher early mortality rate between 300-800 days, while females had a higher mortality rate between 800-1000 days. After more than 1000 days, the mortality rates of female and male mice were similar, indicating that the risk of death is similar at a later stage. These results suggest that genetic loci for lifespan may be age-specific as well as sex-specific.
Therefore, the authors conducted an analysis of four quantitative morphological loci, targeting different sexes and different age stages. The authors found that conditions such as the number of pups and early nutritional acquisition all contribute to longevity. In addition, through the age- and genotype-dependent changes in liver gene expression, the authors obtained prioritized candidate genetic loci.
By jointly analyzing female and male mice, the authors discovered a previously reported gene locus on chromosome 12 [3]. When analyzing the sexes separately, the authors found that females and males have different genetic determinants of longevity. In females, a locus on chromosome 3 is a gene locus associated with longevity, while in males this locus can only be detected when samples that have excluded early deaths are excluded. This suggests that some genetic variants only affect lifespan after a certain age.
To prioritize genes under the longevity locus, the authors analyzed gene expression in the liver of adult and aged mice to look for sex-, age-, and gene-driven expression differences. Interferon-related genes were expressed at higher levels in the liver of female mice, while immune-related genes were overexpressed in the liver of aged mice. Further, the authors assessed genetic regulation of gene expression, most of which were conserved between sex and age. The authors then combined these results with databases from multiple sources from different model organisms and humans to create an interactive resource for prioritizing evolutionarily conserved longevity genes (https://www.systems-genetics.org/itp-longevity).
Longevity-related topics take a long time to study, so the authors decided to verify the longevity genes with higher scores in nematodes with a shorter life history, and identified Hipk1, Ddost, Hspg2, Fgd6, and Pdk1 as candidate longevity-related genes. Among them, Hipk1, Ddost, Hspg2, and Fgd6 will shorten the lifespan of nematodes after knocking out, while Pdk1 will extend lifespan after knocking out (Figure 2).
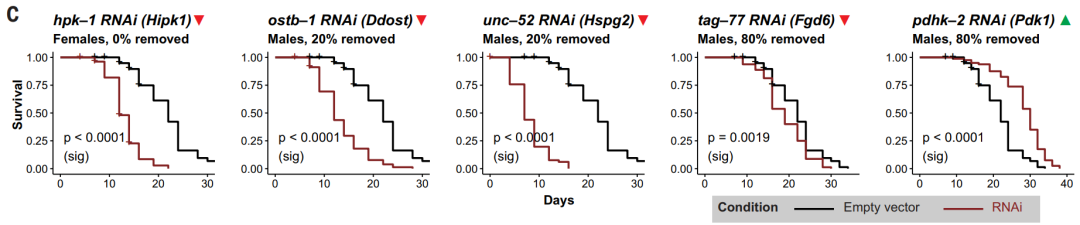
Figure 2 Conserved longevity sites in nematodes
Overall, the study provides insights into the determinants of longevity and finds that genes associated with longevity are associated with gender and different age stages. The collection of this data information constitutes a huge resource database for future research and treatment on aging and longevity.

In the same issue, a viewpoint article was published to comment on it, entitled The genetics of a long life. Healthy aging has been a topic of great concern in recent years. The article highly praised the article for revealing the loci that regulate lifespan in a large-scale comparison of different types of mice and the human genome. Compared with the previous studies on mice, fruit flies, nematodes, etc., more than 2,000 possible longevity gene loci were obtained, which greatly improved the feasibility and detailed conditions of the research and provided valuable resources for further research.
Original link:
https://doi.org/10.1126/science.abo3191